Abstract
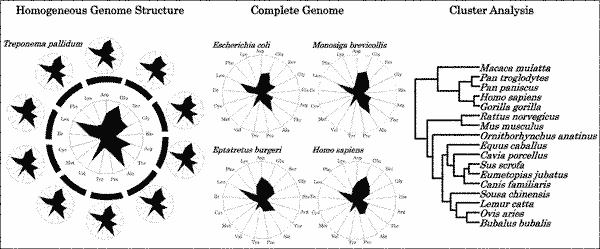
Normalized nucleotide and amino acid contents of complete genome sequences can be visualized as radar charts. The shapes of these charts depict the characteristics of an organism’s genome. The normalized values calculated from the genome sequence theoretically exclude experimental errors. Further, because normalization is independent of both target size and kind, this procedure is applicable not only to single genes but also to whole genomes, which consist of a huge number of different genes. In this review, we discuss the applications of the normalization of the nucleotide and predicted amino acid contents of complete genomes to the investigation of genome structure and to evolutionary research from primitive organisms to Homo sapiens. Some of the results could never have been obtained from the analysis of individual nucleotide or amino acid sequences but were revealed only after the normalization of nucleotide and amino acid contents was applied to genome research. The discovery that genome structure was homogeneous was obtained only after normalization methods were applied to the nucleotide or predicted amino acid contents of genome sequences. Normalization procedures are also applicable to evolutionary research. Thus, normalization of the contents of whole genomes is a useful procedure that can help to characterize organisms.
Keywords: Amino acid composition, Chargaff’s parity rules, Cluster analysis, Evolution, Genome, Normalization, Nucleotide content, Phylogenetic trees.