Abstract
Background: Inhibiting cancer metabolism via glutaminase (GLS) is a promising strategy to disrupt tumor progression. However, the mechanism regarding GLS acetylation remains largely unknown.
Methods: Mitochondrial protein isolation and glutaminase activity assay were used to examine GLS activity. RT-qPCR, western blot, sphere-formation, ALDH activity, and tumor-initiating assays were performed to evaluate the alteration of cell stemness. Co-IP and rescuing experiments were conducted to explore the underlying mechanisms.
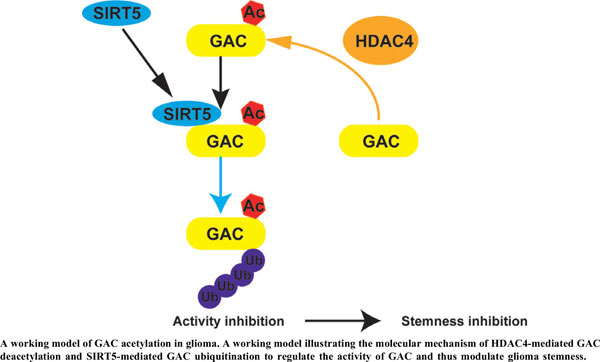
Results: In this study, we demonstrated that GLS acetylation is a vital post-translational modification that inhibits GLS activity in glioma. We identified GLS as deacetylated by HDAC4, a class II deacetylase. GLS acetylation stimulated the interaction between GLS and SIRT5, thereby promoting GLS ubiquitination and inhibiting GLS activity. Furthermore, GLS overexpression suppressed the stemness of glioma cells, which was rescued by the deacetylation of GLS.
Conclusion: Our findings reveal a novel mechanism of GLS regulation by acetylation and ubiquitination that participate in glioma stemness.
Keywords: Glutaminase, cancer metabolism, acetylation, glioma, stemness, mitochondrial protein.
[http://dx.doi.org/10.48095/ccko2022195] [PMID: 35760572]
[http://dx.doi.org/10.1038/s42255-022-00582-0] [PMID: 35760868]
[http://dx.doi.org/10.1158/0008-5472.CAN-13-0308] [PMID: 23824744]
[http://dx.doi.org/10.1126/science.1160809] [PMID: 19460998]
[http://dx.doi.org/10.1016/j.cllc.2013.09.001] [PMID: 24377741]
[http://dx.doi.org/10.1111/cas.15470] [PMID: 35730256]
[http://dx.doi.org/10.1038/s41467-022-31418-w] [PMID: 35764642]
[http://dx.doi.org/10.1016/j.canlet.2019.09.011] [PMID: 31574293]
[http://dx.doi.org/10.4155/fmc-2016-0190] [PMID: 28111979]
[http://dx.doi.org/10.1111/his.14014] [PMID: 31596504]
[http://dx.doi.org/10.3390/cancers13030483] [PMID: 33513833]
[http://dx.doi.org/10.1038/nature07823] [PMID: 19219026]
[http://dx.doi.org/10.1053/j.gastro.2017.12.022] [PMID: 29305935]
[http://dx.doi.org/10.1016/j.cyto.2019.154774] [PMID: 31344597]
[http://dx.doi.org/10.1038/s41422-018-0021-y] [PMID: 29515166]
[http://dx.doi.org/10.1096/fj.202000564R] [PMID: 32519817]
[http://dx.doi.org/10.1002/ctm2.852] [PMID: 35538890]
[http://dx.doi.org/10.1126/science.1175371] [PMID: 19608861]
[http://dx.doi.org/10.1016/j.celrep.2013.07.024] [PMID: 23954790]
[http://dx.doi.org/10.1186/s13045-020-00901-6] [PMID: 32456660]
[http://dx.doi.org/10.1016/j.ebiom.2018.11.063] [PMID: 30555042]
[http://dx.doi.org/10.1016/j.ebiom.2018.12.026] [PMID: 30579869]
[http://dx.doi.org/10.1038/s41598-017-18762-4] [PMID: 29323154]
[http://dx.doi.org/10.1158/0008-5472.CAN-19-3923]
[http://dx.doi.org/10.1038/s41420-020-0258-3] [PMID: 32337072]
[http://dx.doi.org/10.1073/pnas.1911954116] [PMID: 31843902]
[http://dx.doi.org/10.1158/0008-5472.CAN-18-3527] [PMID: 31040157]
[http://dx.doi.org/10.1182/blood-2015-01-621870] [PMID: 26186940]
[http://dx.doi.org/10.1016/j.bbcan.2018.07.007] [PMID: 30053497]
[http://dx.doi.org/10.1016/j.jim.2009.06.008] [PMID: 19567251]
[http://dx.doi.org/10.1007/s00018-021-03818-6] [PMID: 33895866]
[http://dx.doi.org/10.7150/thno.58655] [PMID: 34335968]
[http://dx.doi.org/10.1172/JCI69600] [PMID: 23999442]
[http://dx.doi.org/10.1073/pnas.1003428107] [PMID: 20421486]
[http://dx.doi.org/10.2174/0929867326666190416165004] [PMID: 31038055]
[http://dx.doi.org/10.1038/s41416-022-01805-7] [PMID: 35637410]
[http://dx.doi.org/10.1038/s41419-021-03417-0] [PMID: 33542203]
[http://dx.doi.org/10.18632/aging.100980] [PMID: 27295551]
[http://dx.doi.org/10.7150/ijbs.69882] [PMID: 35864951]
[http://dx.doi.org/10.3233/CBM-182197] [PMID: 30909186]
[http://dx.doi.org/10.1016/j.molcel.2015.10.017] [PMID: 26585387]
[http://dx.doi.org/10.1080/15384101.2022.2040282] [PMID: 35230909]