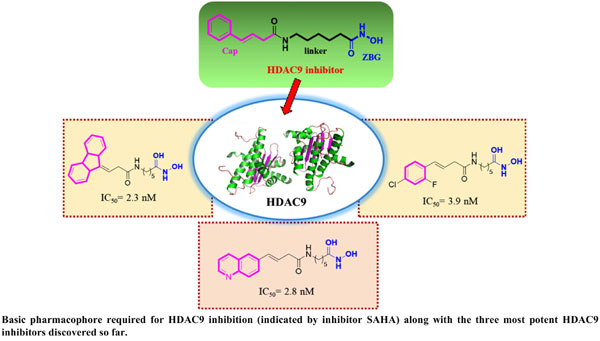
Abstract
HDAC9 is a histone deacetylase enzyme belonging to the class IIa of HDACs which catalyses histone deacetylation. HDAC9 inhibit cell proliferation by repairing DNA, arresting the cell cycle, inducing apoptosis, and altering genetic expression. HDAC9 plays a significant part in human physiological system and are involved in various type of diseases like cancer, diabetes, atherosclerosis and CVD, autoimmune response, inflammatory disease, osteoporosis and liver fibrosis. This review discusses the role of HDAC9 in different diseases and structure-activity relationships (SARs) of various hydroxamate and non-hydroxamate-based inhibitors. SAR of compounds containing several scaffolds have been discussed in detail. Moreover, structural requirements regarding the various components of HDAC9 inhibitor (cap group, linker and zinc-binding group) has been highlighted in this review. Though, HDAC9 is a promising target for the treatment of a number of diseases including cancer, a very few research are available. Thus, this review may provide useful information for designing novel HDAC9 inhibitors to fight against different diseases in the future.
Keywords: Epigenetic, cancer, HDAC9 inhibitor, selectivity, structure-activity relationships, SARs.
[http://dx.doi.org/10.1016/j.ejmech.2014.09.024] [PMID: 25218912]
[http://dx.doi.org/10.2741/2745] [PMID: 17981613]
[http://dx.doi.org/10.1093/jnci/92.15.1210] [PMID: 10922406]
[http://dx.doi.org/10.3390/cancers12061664] [PMID: 32585896]
[http://dx.doi.org/10.1002/(SICI)1097-4652(200007)184:1<1:AID-JCP1>3.0.CO;2-7] [PMID: 10825229]
[http://dx.doi.org/10.1073/pnas.95.7.3519] [PMID: 9520398]
[http://dx.doi.org/10.1007/s13402-021-00626-9] [PMID: 34318404]
[http://dx.doi.org/10.1016/j.molstruc.2022.133967]
[http://dx.doi.org/10.1016/j.molstruc.2022.132833]
[http://dx.doi.org/10.1016/j.jmgm.2023.108510] [PMID: 37216830]
[http://dx.doi.org/10.3390/life11020090] [PMID: 33513699]
[http://dx.doi.org/10.2174/1389557522666220330144151] [PMID: 35362374]
[http://dx.doi.org/10.1111/febs.13061] [PMID: 25244360]
[http://dx.doi.org/10.1093/nar/gks189] [PMID: 22396528]
[http://dx.doi.org/10.1073/pnas.191375098] [PMID: 11535832]
[http://dx.doi.org/10.1021/np200324x] [PMID: 21793558]
[http://dx.doi.org/10.1016/j.jmb.2004.10.033] [PMID: 15567413]
[http://dx.doi.org/10.1080/07391102.2020.1798812] [PMID: 32715940]
[http://dx.doi.org/10.1074/jbc.M110.179333] [PMID: 20947501]
[http://dx.doi.org/10.1073/pnas.97.3.1056] [PMID: 10655483]
[http://dx.doi.org/10.1016/S0167-4781(00)00191-3] [PMID: 11018260]
[http://dx.doi.org/10.1038/s41419-018-0480-6] [PMID: 29691366]
[http://dx.doi.org/10.1038/ni.3464] [PMID: 27240213]
[http://dx.doi.org/10.1007/s11010-016-2690-5]
[http://dx.doi.org/10.1136/jitc-2020-000529] [PMID: 32554611]
[http://dx.doi.org/10.1038/ng.3913] [PMID: 28714975]
[http://dx.doi.org/10.1371/journal.pone.0139262] [PMID: 26488411]
[http://dx.doi.org/10.1038/s41591-019-0492-5] [PMID: 31285632]
[http://dx.doi.org/10.1038/s41588-019-0514-8] [PMID: 31659325]
[http://dx.doi.org/10.1161/STROKEAHA.119.026112] [PMID: 31500558]
[http://dx.doi.org/10.1038/s41569-019-0308-9] [PMID: 31712770]
[http://dx.doi.org/10.1007/s12253-015-9978-8] [PMID: 26347468]
[http://dx.doi.org/10.1161/STROKEAHA.111.000217] [PMID: 23449258]
[http://dx.doi.org/10.1007/s12975-018-0619-x] [PMID: 29651704]
[http://dx.doi.org/10.1016/j.vph.2020.106775] [PMID: 32702412]
[http://dx.doi.org/10.1074/jbc.M111.262964] [PMID: 21680747]
[http://dx.doi.org/10.3390/cells11172698] [PMID: 36078104]
[http://dx.doi.org/10.1111/j.1460-9568.2010.07218.x] [PMID: 20525066]
[http://dx.doi.org/10.1158/1078-0432.CCR-10-0395] [PMID: 20413433]
[http://dx.doi.org/10.18632/oncotarget.3223] [PMID: 25760078]
[http://dx.doi.org/10.1016/j.bbrc.2016.03.129] [PMID: 27033599]
[http://dx.doi.org/10.3390/cancers11101587] [PMID: 31635225]
[http://dx.doi.org/10.3390/cancers12102734] [PMID: 32977608]
[http://dx.doi.org/10.3390/biomedicines10020374] [PMID: 35203583]
[http://dx.doi.org/10.3892/mmr.2019.10869] [PMID: 31974610]
[http://dx.doi.org/10.1038/s12276-019-0301-8] [PMID: 31451695]
[http://dx.doi.org/10.18632/oncotarget.7564] [PMID: 26930713]
[http://dx.doi.org/10.2147/OTT.S164583] [PMID: 29713186]
[http://dx.doi.org/10.1016/j.bbrc.2018.06.120] [PMID: 29936177]
[http://dx.doi.org/10.7150/thno.44997] [PMID: 33042272]
[http://dx.doi.org/10.1007/s00280-019-04024-9] [PMID: 31907648]
[http://dx.doi.org/10.1053/j.gastro.2009.10.037] [PMID: 19879272]
[http://dx.doi.org/10.2337/db11-0440] [PMID: 21953612]
[http://dx.doi.org/10.1242/dev.125674] [PMID: 26483211]
[http://dx.doi.org/10.1161/STROKEAHA.113.002707] [PMID: 24262325]
[http://dx.doi.org/10.1016/j.bbrc.2018.07.043] [PMID: 30031609]
[http://dx.doi.org/10.1111/ane.12847] [PMID: 28975602]
[http://dx.doi.org/10.1038/srep41538] [PMID: 28145521]
[http://dx.doi.org/10.4093/dmj.2019.0243] [PMID: 32347025]
[http://dx.doi.org/10.1038/nrdp.2015.56] [PMID: 27188934]
[http://dx.doi.org/10.1007/s12264-019-00361-0] [PMID: 30887246]
[http://dx.doi.org/10.1161/CIRCRESAHA.120.316743] [PMID: 32546048]
[http://dx.doi.org/10.1161/STROKEAHA.114.007213] [PMID: 25388417]
[http://dx.doi.org/10.1161/CIRCRESAHA.120.317723] [PMID: 32853095]
[http://dx.doi.org/10.1161/ATVBAHA.114.303393] [PMID: 25035344]
[http://dx.doi.org/10.1016/S0092-8674(02)00861-9] [PMID: 12202037]
[http://dx.doi.org/10.1016/j.cell.2008.12.040] [PMID: 19303849]
[http://dx.doi.org/10.4161/adip.28814] [PMID: 26317058]
[http://dx.doi.org/10.2174/1389557521666211116122232] [PMID: 34784862]
[http://dx.doi.org/10.2174/1389557521666211116123002] [PMID: 34784864]
[http://dx.doi.org/10.1038/s41598-017-06328-3] [PMID: 28733598]
[http://dx.doi.org/10.1074/jbc.M115.681627] [PMID: 26504089]
[http://dx.doi.org/10.2337/db15-0197] [PMID: 26420860]
[http://dx.doi.org/10.1172/JCI77716] [PMID: 25751060]
[http://dx.doi.org/10.1186/s13148-020-00858-w] [PMID: 32410704]
[http://dx.doi.org/10.1038/s41467-023-38771-4] [PMID: 37230975]
[http://dx.doi.org/10.1038/sj.ki.5000054] [PMID: 16408108]
[http://dx.doi.org/10.1080/15548627.2016.1190071] [PMID: 27304991]
[http://dx.doi.org/10.1016/j.kint.2022.03.022] [PMID: 35483529]
[http://dx.doi.org/10.1016/j.stem.2008.01.014] [PMID: 18371453]
[http://dx.doi.org/10.1016/j.kint.2021.09.027] [PMID: 34757120]
[http://dx.doi.org/10.1002/cphy.c120035] [PMID: 24265236]
[http://dx.doi.org/10.1016/j.jnutbio.2016.11.003] [PMID: 27915160]
[http://dx.doi.org/10.3390/cells9102321] [PMID: 33086678]
[http://dx.doi.org/10.1016/j.jcmgh.2017.04.007] [PMID: 28593184]
[http://dx.doi.org/10.1053/j.gastro.2008.03.003] [PMID: 18471545]
[http://dx.doi.org/10.1053/j.gastro.2004.08.028] [PMID: 15481004]
[http://dx.doi.org/10.1016/j.taap.2016.07.003] [PMID: 27396813]
[http://dx.doi.org/10.3390/ijms20102507] [PMID: 31117267]
[http://dx.doi.org/10.1177/1535370215584933] [PMID: 25966982]
[http://dx.doi.org/10.1152/physrev.00013.2007] [PMID: 18195085]
[http://dx.doi.org/10.1371/journal.pone.0055786] [PMID: 23383282]
[http://dx.doi.org/10.1016/j.omtn.2021.08.031] [PMID: 34976438]
[http://dx.doi.org/10.1007/s11033-020-05431-5] [PMID: 32277440]
[http://dx.doi.org/10.1074/jbc.M111.262964] [PMID: 21680747]
[http://dx.doi.org/10.2337/db13-1148] [PMID: 24101673]
[http://dx.doi.org/10.1038/onc.2008.25] [PMID: 18391974]
[http://dx.doi.org/10.1186/s13287-020-01785-6] [PMID: 32620134]
[http://dx.doi.org/10.2147/CIA.S361008] [PMID: 35592642]
[http://dx.doi.org/10.1210/me.2014-1365] [PMID: 25793404]
[http://dx.doi.org/10.1155/2016/1652417] [PMID: 27073801]
[http://dx.doi.org/10.1074/jbc.M111.233932] [PMID: 21708950]
[http://dx.doi.org/10.1016/j.coi.2010.08.013] [PMID: 20869864]
[http://dx.doi.org/10.1126/sciimmunol.aah4609] [PMID: 28783689]
[http://dx.doi.org/10.4049/jimmunol.1800893] [PMID: 30737272]
[http://dx.doi.org/10.1128/MCB.01856-08] [PMID: 19414603]
[http://dx.doi.org/10.1038/nm1652] [PMID: 17922010]
[http://dx.doi.org/10.2174/1389557519666211221144013] [PMID: 34939540]
[http://dx.doi.org/10.1016/j.bcp.2022.115301] [PMID: 36265594]
[http://dx.doi.org/10.1016/j.ejmech.2023.115594] [PMID: 37429084]
[http://dx.doi.org/10.1080/07391102.2023.2227710]
[http://dx.doi.org/10.1016/j.febslet.2012.05.054] [PMID: 22684007]
[http://dx.doi.org/10.1159/000369733] [PMID: 25613642]
[http://dx.doi.org/10.1021/ml500024s] [PMID: 24900884]
[http://dx.doi.org/10.1016/j.bmc.2014.05.001] [PMID: 24864038]
[http://dx.doi.org/10.1021/jm101092u] [PMID: 21073160]
[http://dx.doi.org/10.1016/j.ejmech.2008.06.020] [PMID: 18672316]
[http://dx.doi.org/10.1021/acs.jmedchem.9b02178] [PMID: 32153186]
[http://dx.doi.org/10.1016/j.ejmech.2019.05.089] [PMID: 31177073]
[http://dx.doi.org/10.1016/j.ejmech.2017.11.092] [PMID: 29223096]
[http://dx.doi.org/10.1021/acs.jmedchem.6b00579] [PMID: 27186676]
[http://dx.doi.org/10.1016/j.bmcl.2018.11.009] [PMID: 30463802]
[http://dx.doi.org/10.1021/jm401899x] [PMID: 24766560]
[http://dx.doi.org/10.1021/ml300371t] [PMID: 24900575]
[http://dx.doi.org/10.1021/ml1001954] [PMID: 21874153]
[http://dx.doi.org/10.1021/acs.jmedchem.5b01342] [PMID: 26443078]
[http://dx.doi.org/10.1021/acs.jmedchem.7b01404] [PMID: 29304284]
[http://dx.doi.org/10.1016/j.ejmech.2019.111725] [PMID: 31655430]
[http://dx.doi.org/10.1021/acs.jmedchem.5b01044] [PMID: 26331334]